IntroductionIn this post I am going to show how to extract data from web pages in table format, transform these data into spatial objects in R and then plot them in maps.
ProcedureFor this project we need the following two packages: XML and raster.
The first package is used to extract data from HTML pages, in particular from the sections marked with the tag <table>, which marks text ordered in a table format on HTML pages.
The example below is created using a sample of code from: http://www.w3schools.com/Html/html_tables.asp. If we look at the code in plain text you can see that is included within the tag <table></table>.
<table style="width:100%">
<tr>
<td>Jill</td>
<td>Smith</td>
<td>50</td>
</tr>
<tr>
<td>Eve</td>
<td>Jackson</td>
<td>94</td>
</tr>
<tr>
<td>John</td>
<td>Doe</td>
<td>80</td>
</tr>
</table>
In an HTML page the code above would look like this:
| Jill | Smith | 50 |
| Eve | Jackson | 94 |
| John | Doe | 80 |
In the package XML we have a function, readHTMLTable, which is able to extract only the data written within the two tags
<table> and
</table>. Therefore we can use it to download data potentially from every web page.
The problem is the data are imported in R in textual string and therefor they need some processing before we can actually use them.
Before starting importing data from the web however, we are going to download and import a shapefile with the borders of all the countries in the world from this page:
thematicmapping.orgThe code for doing it is the following:
download.file("http://thematicmapping.org/downloads/TM_WORLD_BORDERS_SIMPL-0.3.zip",destfile="TM_WORLD_BORDERS_SIMPL-0.3.zip")
unzip("TM_WORLD_BORDERS_SIMPL-0.3.zip",exdir=getwd())
polygons <- shapefile("TM_WORLD_BORDERS_SIMPL-0.3.shp")In the first line we use the function download.file to download the zip containing the shapefile. Here we need to specify the destination, which I called with the same name of the object.
At this point we have downloaded a zip file in the working directory, so we need to extract its contents. We can do that using unzip, which require as arguments the name of the zip file and the directory where to extract its contents, in this case I used
getwd() to extract everything in the working directory.
Finally we can open the shapefile using the function with the same name, creating the object polygons, which looks like this:
> polygons
class : SpatialPolygonsDataFrame
features : 246
extent : -180,180, -90,83.57027(xmin, xmax, ymin, ymax)
coord. ref. : +proj=longlat +datum=WGS84 +no_defs
variables : 11
names : FIPS, ISO2, ISO3, UN, NAME, AREA, POP2005, REGION, SUBREGION, LON, LAT
min values : AA, AD, ABW,4,Ã…land Islands,0,0,0,0, -102.535, -10.444
max values : ZI, ZW, ZWE,894, Zimbabwe,1638094,1312978855,150,155,179.219,78.830
As you can see from the column
NAME, some country names are not well read in R and this is something that we would need to correct later on in the exercise.
At this point we can start downloading economic data from the web pages of The World Bank. An example of the type of data and the page we are going to query is here:
http://data.worldbank.org/indicator/EN.ATM.CO2E.PCFor this exrcise we are going to download the following data: CO2 emissions (metric tons per capita), Population in urban agglomerations of more than 1 million, Population density (people per sq. km of land area), Population in largest city, GDP per capita (current US$), GDP (current US$), Adjusted net national income per capita (current US$), Adjusted net national income (current US$), Electric power consumption (kWh per capita), Electric power consumption (kWh), Electricity production (kWh).
The line of code to import this page into R is the following:
CO2_emissions_Tons.per.Capita_HTML <- readHTMLTable("http://data.worldbank.org/indicator/EN.ATM.CO2E.PC")The object
CO2_emissions_Tons.per.Capita_HTML is a list containing two elements:
> str(CO2_emissions_Tons.per.Capita_HTML)
List of 2
$ NULL:'data.frame': 213 obs. of 4 variables:
..$
Country name : Factor w/ 213levels"Afghanistan",..: 12345678910 ...
..$
2010 : Factor w/ 93levels"","0.0","0.1",..: 516521771773631550 ...
..$
: Factor w/ 1 level "": 1111111111 ...
..$
: Factor w/ 1 level "": 1111111111 ...
$ NULL:'data.frame': 10 obs. of 2 variables:
..$ V1: Factor w/ 10levels"Agriculture & Rural Development",..: 12345678910
..$ V2: Factor w/ 10levels"Health","Infrastructure",..: 12345678910
That is because at the very end of the page there is a list of topics that is also displayed using the tag
<table>. For this reason if we want to import only the first table we need to subset the object like so:
CO2_emissions_Tons.per.Capita_HTML <- readHTMLTable("http://data.worldbank.org/indicator/EN.ATM.CO2E.PC")[[1]]In yellow is highlighted the subsetting call.
The object
CO2_emissions_Tons.per.Capita_HTML is a
data.frame:
> str(CO2_emissions_Tons.per.Capita_HTML)
'data.frame': 213 obs. of 4 variables:
$
Country name : Factor w/ 213levels"Afghanistan",..: 12345678910 ...
$
2010 : Factor w/ 93levels"","0.0","0.1",..: 516521771773631550 ...
$
: Factor w/ 1 level "": 1111111111 ...
$
: Factor w/ 1 level "": 1111111111 ...
It has 4 columns, two of which are empty; we only nee the column named "Country name" and the one with data for the year 2010. The problem is, not every dataset has the data from the same years; in some case we only have 2010, like here, in other we have more than one year. Therefore, depending on what we want to achieve the approach to do it would be slightly different. For example, if we just want to plot the most recent data we could subset each dataset keeping only the most columns with the most recent year and exclude the others. Alternatively, if we need data from a particular year we could create variable and then try to mach it to the column in each dataset.
I will try to show how to achieve both.
The first example finds the most recent year in the data and extracts only those values.
To do this we just need to find the highest number among the column names, with the following line:
This lines does several thing at once; first uses the function
names() to extract the names of the columns in the
data.frame, in textual format. Then converts these texts into numbers, when possible (for example the column named "Country name" would return NA), with the function
as.numeric(), which forces the conversion. At this point we have a list of numbers and NAs, from which we can calculate the maximum using the function
max() with the option
na.rm=T that excludes the NAs for the computation. The
as.numeric() call returns a warning, since it creates NAs, but you should not worry about it.
The result for CO2 emission is a single number: 2010
Once we have identified the most recent year in the dataset, we can identify its column in the
data.frame. We have two approaches to do so, the first is based on the function
which():
This returns a warning, for the same reason explained above, but it does the job. Another possibility is to use the function
grep(), which is able to search within character vectors for particular patterns.
We can use this function like so:
CO2.col <- grep(x=names(CO2_emissions_Tons.per.Capita_HTML),pattern=paste(CO2.year))
This function takes two argument: x and pattern.
The first is the character vector where to search for particular words, the second is the pattern to be identified in x. In this case we are taking the names of the columns and we are trying to identify the one string that contains the number 2010, which needs to be in a character format, hence the use of
paste(). Both functions return the number of the element corresponding to the clause in the character vector. In other words, if we have for example a character vector composed by the elements: "Banana", "Pear", "Apple" and we apply these two function searching for the word "Pear", they will return the number 2, which is position of the word in the vector.
A second way of extracting data from the economic tables is by defining a particular year and then match it to the data. For this we first need to create a new variable:
The object
CO2.year would then be equal to the value assigned to the variable
YEAR:
The variable
YEAR can also be used to identify the column in the data.frame:
CO2.col <- grep(x=names(CO2_emissions_Tons.per.Capita_HTML),pattern=paste(YEAR))
At this point we have all we need to attach the economic data to the polygon shapefile, matching the Country names in the economic tables with the names in the shapefile. As I mentioned before, some names do not match between the two datasets and in the case of "Western Sahara" the state is not present in the economic table. For this reason, before we can proceed in creating the map we need to modify the polygons object to match the economic table using the following code:
#Some Country names need to be changed for inconsistencies between datasets
polygons[polygons$NAME=="Libyan Arab Jamahiriya","NAME"]<- "Libya"
polygons[polygons$NAME=="Democratic Republic of the Congo","NAME"]<- "Congo, Dem. Rep."
polygons[polygons$NAME=="Congo","NAME"]<- "Congo, Rep."
polygons[polygons$NAME=="Kyrgyzstan","NAME"]<- "Kyrgyz Republic"
polygons[polygons$NAME=="United Republic of Tanzania","NAME"]<- "Tanzania"
polygons[polygons$NAME=="Iran (Islamic Republic of)","NAME"]<- "Iran, Islamic Rep."
#The States of "Western Sahara", "French Guyana" do not exist in the World Bank Database
These are the names that we have to replace manually, otherwise there would be no way to make R understand that these names refer to the same states. However, even after these changes we cannot just link the list of countries of the shapefile with the one on the World Bank data; there are other names for which there is no direct correlation. For example, on the World Bank website The Bahamas is referred to as "Bahamas, The", while in the polygon shapefile the same country is referred to as simply "Bahamas". If we try to match the two lists we would obtain an NA. However, for such a case we can use again the function
grep() to identify, in the list of countries on the World Bank, the one string than contain the word "Bahamas", using a line of code like the following:
This code extracts from the World Bank data the CO2 value, for the year that we defined earlier, related to the country defined in the polygon shapefile, for a certain row, which here is defined simply as "row". This line is again rich of nested functions. The innermost is
grep():
grep(x=paste(CO2_emissions_Tons.per.Capita_HTML[,1]),pattern=paste(polygons[row,]$NAME))
with this line we are searching in the list of countries the one word that corresponds to the name of the country in the polygons shapefile. As mentioned before the function
grep() return a number, which identify the element in the vector. We can use this number to extract, from the object
CO2_emissions_Tons.per.Capita_HTML the corresponding line, and the column corresponding to the value in the object
CO2.col (that we created above).
This line works in the case of the Bahamas, because there is only one element in the list of countries of the World Bank with than word. However, there are other cases where more than multiple countries have the same word in their name, for example: "China", "Hong Kong SAR, China" and "Macao SAR, China". If we use the line above with China for example, since the function
grep() looks for word that contain China, it would identify all of them and it would be tricky in R to automatically identify the correct element. To resolve to such a situation we can add another line specifically created for these cases:
polygons[row,"CO2"]<- as.numeric(paste(CO2_emissions_Tons.per.Capita_HTML[paste(CO2_emissions_Tons.per.Capita_HTML[,1])==paste(polygons[row,]$NAME),CO2.col]))
In this case we have to match the exact words so we can just use a == clause and extract only the element that corresponds to the word "China". The other two countries, Hong Kong and Macao, would be recognized by the grep function by pattern recognition.
Now that we defined all the rules we would nee we can create a loop to automatically extract all the values of CO2 for each country in the World Bank and attach them to the polygon shapefile:
polygons$CO2 <- c()
for(rowin1:length(polygons)){
if(any(grep(x=paste(CO2_emissions_Tons.per.Capita_HTML[,1]),pattern=paste(polygons[row,]$NAME)))&length(grep(x=paste(CO2_emissions_Tons.per.Capita_HTML[,1]),pattern=paste(polygons[row,]$NAME)))==1){
polygons[row,"CO2"]<- as.numeric(paste(CO2_emissions_Tons.per.Capita_HTML[grep(x=paste(CO2_emissions_Tons.per.Capita_HTML[,1]),pattern=paste(polygons[row,]$NAME)),CO2.col]))
}
if(any(grep(x=paste(CO2_emissions_Tons.per.Capita_HTML[,1]),pattern=paste(polygons[row,]$NAME)))&length(grep(x=paste(CO2_emissions_Tons.per.Capita_HTML[,1]),pattern=paste(polygons[row,]$NAME)))>1){
polygons[row,"CO2"]<- as.numeric(paste(CO2_emissions_Tons.per.Capita_HTML[paste(CO2_emissions_Tons.per.Capita_HTML[,1])==paste(polygons[row,]$NAME),CO2.col]))
}
}
First of all we create a new column in the object polygons named CO2, which for the time being is an empty vector. The we start the loop that iterates from 1 to the number of rows of the object polygons, for each row it needs to extract the CO2 values that corresponds to the country.
Since we have the two situations described above with the country names, we need to define two if sentences to guide the looping and avoid errors. We use two functions for this:
any() and
length().
The function
any() examines the output of the
grep() function and returns
TRUE only if
grep() returns a number. If
grep() does not find any country in the World Bank data that corresponds to the country in the polygons shapefile the loop will return an NA, because both if clauses would return
FALSE. The function length() is used to discriminate between the two situations we described above. i.e. we have one line for situations in which
grep() returns just one number and another when
grep() identify multiple countries with the same pattern.
Once we finished the loop the object polygons has a column names CO2, which we can use to plot the map of the CO2 Emission using the function
spplot():
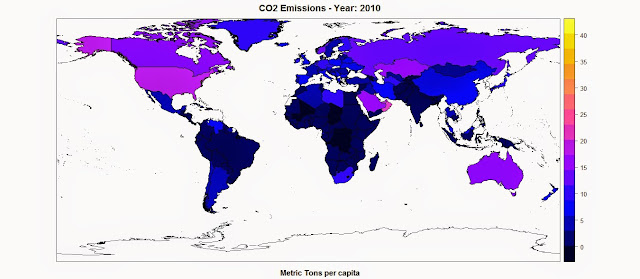
spplot(polygons,"CO2",main=paste("CO2 Emissions - Year:",CO2.year),sub="Metric Tons per capita")The result is the following map, where the function spplot creates also a color scale from the data:
R code snippets created by Pretty R at inside-R.orgNow we can apply the same exact approach for the other datasets we need to download from the World Bank and create maps of all the economic indexes available. The full script is available below (it was fully tested in date 12th May 2015).
library(raster)
library(XML)
library(rgdal)
#Change the following line to set the working directory
setwd("...")
#Download, unzip and load the polygon shapefile with the countries' borders
download.file("http://thematicmapping.org/downloads/TM_WORLD_BORDERS_SIMPL-0.3.zip",destfile="TM_WORLD_BORDERS_SIMPL-0.3.zip")
unzip("TM_WORLD_BORDERS_SIMPL-0.3.zip",exdir=getwd())
polygons <- shapefile("TM_WORLD_BORDERS_SIMPL-0.3.shp")
#Read Economic data tables from the World Bank website
CO2_emissions_Tons.per.Capita_HTML <- readHTMLTable("http://data.worldbank.org/indicator/EN.ATM.CO2E.PC")[[1]]
Population_urban.more.1.mln_HTML <- readHTMLTable("http://data.worldbank.org/indicator/EN.URB.MCTY")[[1]]
Population_Density_HTML <- readHTMLTable("http://data.worldbank.org/indicator/EN.POP.DNST")[[1]]
Population_Largest_Cities_HTML <- readHTMLTable("http://data.worldbank.org/indicator/EN.URB.LCTY")[[1]]
GDP_capita_HTML <- readHTMLTable("http://data.worldbank.org/indicator/NY.GDP.PCAP.CD")[[1]]
GDP_HTML <- readHTMLTable("http://data.worldbank.org/indicator/NY.GDP.MKTP.CD")[[1]]
Adj_Income_capita_HTML <- readHTMLTable("http://data.worldbank.org/indicator/NY.ADJ.NNTY.PC.CD")[[1]]
Adj_Income_HTML <- readHTMLTable("http://data.worldbank.org/indicator/NY.ADJ.NNTY.CD")[[1]]
Elect_Consumption_capita_HTML <- readHTMLTable("http://data.worldbank.org/indicator/EG.USE.ELEC.KH.PC")[[1]]
Elect_Consumption_HTML <- readHTMLTable("http://data.worldbank.org/indicator/EG.USE.ELEC.KH")[[1]]
Elect_Production_HTML <- readHTMLTable("http://data.worldbank.org/indicator/EG.ELC.PROD.KH")[[1]]
#First Approach - Find the most recent data
#Maximum Year
CO2.year <- max(as.numeric(names(CO2_emissions_Tons.per.Capita_HTML)),na.rm=T)
PoplUrb.year <- max(as.numeric(names(Population_urban.more.1.mln_HTML)),na.rm=T)
PoplDens.year <- max(as.numeric(names(Population_Density_HTML)),na.rm=T)
PoplLarg.year <- max(as.numeric(names(Population_Largest_Cities_HTML)),na.rm=T)
GDPcap.year <- max(as.numeric(names(GDP_capita_HTML)),na.rm=T)
GDP.year <- max(as.numeric(names(GDP_HTML)),na.rm=T)
AdjInc.cap.year <- max(as.numeric(names(Adj_Income_capita_HTML)),na.rm=T)
AdjInc.year <- max(as.numeric(names(Adj_Income_HTML)),na.rm=T)
EleCon.cap.year <- max(as.numeric(names(Elect_Consumption_capita_HTML)),na.rm=T)
EleCon.year <- max(as.numeric(names(Elect_Consumption_HTML)),na.rm=T)
ElecProd.year <- max(as.numeric(names(Elect_Production_HTML)),na.rm=T)
#Column Maximum Year
CO2.col <- grep(x=names(CO2_emissions_Tons.per.Capita_HTML),pattern=paste(CO2.year))
PoplUrb.col <- grep(x=names(Population_urban.more.1.mln_HTML),pattern=paste(PoplUrb.year))
PoplDens.col <- grep(x=names(Population_Density_HTML),pattern=paste(PoplDens.year))
PoplLarg.col <- grep(x=names(Population_Largest_Cities_HTML),pattern=paste(PoplLarg.year))
GDPcap.col <- grep(x=names(GDP_capita_HTML),pattern=paste(GDPcap.year))
GDP.col <- grep(x=names(GDP_HTML),pattern=paste(GDP.year))
AdjInc.cap.col <- grep(x=names(Adj_Income_capita_HTML),pattern=paste(AdjInc.cap.year))
AdjInc.col <- grep(x=names(Adj_Income_HTML),pattern=paste(AdjInc.year))
EleCon.cap.col <- grep(x=names(Elect_Consumption_capita_HTML),pattern=paste(EleCon.cap.year))
EleCon.col <- grep(x=names(Elect_Consumption_HTML),pattern=paste(EleCon.year))
ElecProd.col <- grep(x=names(Elect_Production_HTML),pattern=paste(ElecProd.year))
#Second Approach - Find data for specific Years
YEAR = 2010
#Year
CO2.year <- YEAR
PoplUrb.year <- YEAR
PoplDens.year <- YEAR
PoplLarg.year <- YEAR
GDPcap.year <- YEAR
GDP.year <- YEAR
AdjInc.cap.year <- YEAR
AdjInc.year <- YEAR
EleCon.cap.year <- YEAR
EleCon.year <- YEAR
ElecProd.year <- YEAR
#Column Maximum Year
CO2.col <- grep(x=names(CO2_emissions_Tons.per.Capita_HTML),pattern=paste(YEAR))
PoplUrb.col <- grep(x=names(Population_urban.more.1.mln_HTML),pattern=paste(YEAR))
PoplDens.col <- grep(x=names(Population_Density_HTML),pattern=paste(YEAR))
PoplLarg.col <- grep(x=names(Population_Largest_Cities_HTML),pattern=paste(YEAR))
GDPcap.col <- grep(x=names(GDP_capita_HTML),pattern=paste(YEAR))
GDP.col <- grep(x=names(GDP_HTML),pattern=paste(YEAR))
AdjInc.cap.col <- grep(x=names(Adj_Income_capita_HTML),pattern=paste(YEAR))
AdjInc.col <- grep(x=names(Adj_Income_HTML),pattern=paste(YEAR))
EleCon.cap.col <- grep(x=names(Elect_Consumption_capita_HTML),pattern=paste(YEAR))
EleCon.col <- grep(x=names(Elect_Consumption_HTML),pattern=paste(YEAR))
ElecProd.col <- grep(x=names(Elect_Production_HTML),pattern=paste(YEAR))
#Some Country names need to be changed for inconsistencies between datasets
polygons[polygons$NAME=="Libyan Arab Jamahiriya","NAME"] <- "Libya"
polygons[polygons$NAME=="Democratic Republic of the Congo","NAME"] <- "Congo, Dem. Rep."
polygons[polygons$NAME=="Congo","NAME"] <- "Congo, Rep."
polygons[polygons$NAME=="Kyrgyzstan","NAME"] <- "Kyrgyz Republic"
polygons[polygons$NAME=="United Republic of Tanzania","NAME"] <- "Tanzania"
polygons[polygons$NAME=="Iran (Islamic Republic of)","NAME"] <- "Iran, Islamic Rep."
#The States of "Western Sahara", "French Guyana" do not exist in the World Bank Database
#Now we can start the loop to add the economic data to the polygon shapefile
polygons$CO2 <- c()
polygons$PoplUrb <- c()
polygons$PoplDens <- c()
polygons$PoplLargCit <- c()
polygons$GDP.capita <- c()
polygons$GDP <- c()
polygons$AdjInc.capita <- c()
polygons$AdjInc <- c()
polygons$ElectConsumpt.capita <- c()
polygons$ElectConsumpt <- c()
polygons$ElectProduct <- c()
for(row in 1:length(polygons)){
if(any(grep(x=paste(CO2_emissions_Tons.per.Capita_HTML[,1]),pattern=paste(polygons[row,]$NAME)))&length(grep(x=paste(CO2_emissions_Tons.per.Capita_HTML[,1]),pattern=paste(polygons[row,]$NAME)))==1){
polygons[row,"CO2"] <- as.numeric(paste(CO2_emissions_Tons.per.Capita_HTML[grep(x=paste(CO2_emissions_Tons.per.Capita_HTML[,1]),pattern=paste(polygons[row,]$NAME)),CO2.col]))
polygons[row,"PoplUrb"] <- as.numeric(gsub(",","",paste(Population_urban.more.1.mln_HTML[grep(x=paste(Population_urban.more.1.mln_HTML[,1]),pattern=paste(polygons[row,]$NAME)),PoplUrb.col])))
polygons[row,"PoplDens"] <- as.numeric(paste(Population_Density_HTML[grep(x=paste(Population_Density_HTML[,1]),pattern=paste(polygons[row,]$NAME)),PoplDens.col]))
polygons[row,"PoplLargCit"] <- as.numeric(gsub(",","",paste(Population_Largest_Cities_HTML[grep(x=paste(Population_Largest_Cities_HTML[,1]),pattern=paste(polygons[row,]$NAME)),PoplLarg.col])))
polygons[row,"GDP.capita"] <- as.numeric(gsub(",","",paste(GDP_capita_HTML[grep(x=paste(GDP_capita_HTML[,1]),pattern=paste(polygons[row,]$NAME)),GDPcap.col])))
polygons[row,"GDP"] <- as.numeric(gsub(",","",paste(GDP_HTML[grep(x=paste(GDP_HTML[,1]),pattern=paste(polygons[row,]$NAME)),GDP.col])))
polygons[row,"AdjInc.capita"] <- as.numeric(gsub(",","",paste(Adj_Income_capita_HTML[grep(x=paste(Adj_Income_capita_HTML[,1]),pattern=paste(polygons[row,]$NAME)),AdjInc.cap.col])))
polygons[row,"AdjInc"] <- as.numeric(gsub(",","",paste(Adj_Income_HTML[grep(x=paste(Adj_Income_HTML[,1]),pattern=paste(polygons[row,]$NAME)),AdjInc.col])))
polygons[row,"ElectConsumpt.capita"] <- as.numeric(gsub(",","",paste(Elect_Consumption_capita_HTML[grep(x=paste(Elect_Consumption_capita_HTML[,1]),pattern=paste(polygons[row,]$NAME)),EleCon.cap.col])))
polygons[row,"ElectConsumpt"] <- as.numeric(gsub(",","",paste(Elect_Consumption_HTML[grep(x=paste(Elect_Consumption_HTML[,1]),pattern=paste(polygons[row,]$NAME)),EleCon.col])))
polygons[row,"ElectProduct"] <- as.numeric(gsub(",","",paste(Elect_Production_HTML[grep(x=paste(Elect_Production_HTML[,1]),pattern=paste(polygons[row,]$NAME)),ElecProd.col])))
}
if(any(grep(x=paste(CO2_emissions_Tons.per.Capita_HTML[,1]),pattern=paste(polygons[row,]$NAME)))&length(grep(x=paste(CO2_emissions_Tons.per.Capita_HTML[,1]),pattern=paste(polygons[row,]$NAME)))>1){
polygons[row,"CO2"] <- as.numeric(paste(CO2_emissions_Tons.per.Capita_HTML[paste(CO2_emissions_Tons.per.Capita_HTML[,1])==paste(polygons[row,]$NAME),CO2.col]))
polygons[row,"PoplUrb"] <- as.numeric(gsub(",","",paste(Population_urban.more.1.mln_HTML[paste(Population_urban.more.1.mln_HTML[,1])==paste(polygons[row,]$NAME),PoplUrb.col])))
polygons[row,"PoplDens"] <- as.numeric(paste(Population_Density_HTML[paste(Population_Density_HTML[,1])==paste(polygons[row,]$NAME),PoplDens.col]))
polygons[row,"PoplLargCit"] <- as.numeric(gsub(",","",paste(Population_Largest_Cities_HTML[paste(Population_Largest_Cities_HTML[,1])==paste(polygons[row,]$NAME),PoplLarg.col])))
polygons[row,"GDP.capita"] <- as.numeric(gsub(",","",paste(GDP_capita_HTML[paste(GDP_capita_HTML[,1])==paste(polygons[row,]$NAME),GDPcap.col])))
polygons[row,"GDP"] <- as.numeric(gsub(",","",paste(GDP_HTML[paste(GDP_HTML[,1])==paste(polygons[row,]$NAME),GDP.col])))
polygons[row,"AdjInc.capita"] <- as.numeric(gsub(",","",paste(Adj_Income_capita_HTML[paste(Adj_Income_capita_HTML[,1])==paste(polygons[row,]$NAME),AdjInc.cap.col])))
polygons[row,"AdjInc"] <- as.numeric(gsub(",","",paste(Adj_Income_HTML[paste(Adj_Income_HTML[,1])==paste(polygons[row,]$NAME),AdjInc.col])))
polygons[row,"ElectConsumpt.capita"] <- as.numeric(gsub(",","",paste(Elect_Consumption_capita_HTML[paste(Elect_Consumption_capita_HTML[,1])==paste(polygons[row,]$NAME),EleCon.cap.col])))
polygons[row,"ElectConsumpt"] <- as.numeric(gsub(",","",paste(Elect_Consumption_HTML[paste(Elect_Consumption_HTML[,1])==paste(polygons[row,]$NAME),EleCon.col])))
polygons[row,"ElectProduct"] <- as.numeric(gsub(",","",paste(Elect_Production_HTML[paste(Elect_Production_HTML[,1])==paste(polygons[row,]$NAME),ElecProd.col])))
}
}
#Spatial Plots
spplot(polygons,"CO2",main=paste("CO2 Emissions - Year:",CO2.year),sub="Metric Tons per capita")
spplot(polygons,"PoplUrb",main=paste("Population - Year:",PoplUrb.year),sub="In urban agglomerations of more than 1 million")
spplot(polygons,"PoplDens",main=paste("Population Density - Year:",PoplDens.year),sub="People per sq. km of land area")
spplot(polygons,"PoplLargCit",main=paste("Population in largest city - Year:",PoplLarg.year))
spplot(polygons,"GDP.capita",main=paste("GDP per capita - Year:",GDPcap.year),sub="Currency: USD")
spplot(polygons,"GDP",main=paste("GDP - Year:",GDP.year),sub="Currency: USD")
spplot(polygons,"AdjInc.capita",main=paste("Adjusted net national income per capita - Year:",AdjInc.cap.year),sub="Currency: USD")
spplot(polygons,"AdjInc",main=paste("Adjusted net national income - Year:",AdjInc.year),sub="Currency: USD")
spplot(polygons,"ElectConsumpt.capita",main=paste("Electric power consumption per capita - Year:",EleCon.cap.year),sub="kWh per capita")
spplot(polygons,"ElectConsumpt",main=paste("Electric power consumption - Year:",EleCon.year),sub="kWh")
spplot(polygons,"ElectProduct",main=paste("Electricity production - Year:",ElecProd.year),sub="kWh")